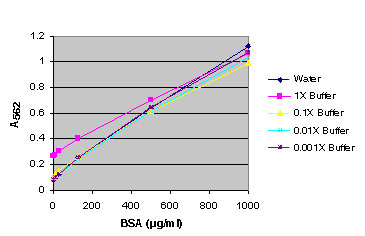
TR0068-Protein-assay-compatibility.pdf - TECH TIP #68 Protein assay compatibility table TR0068.5 Introduction No single protein assay method is | Course Hero

TR0068-Protein-assay-compatibility.pdf - TECH TIP #68 Protein assay compatibility table TR0068.5 Introduction No single protein assay method is | Course Hero

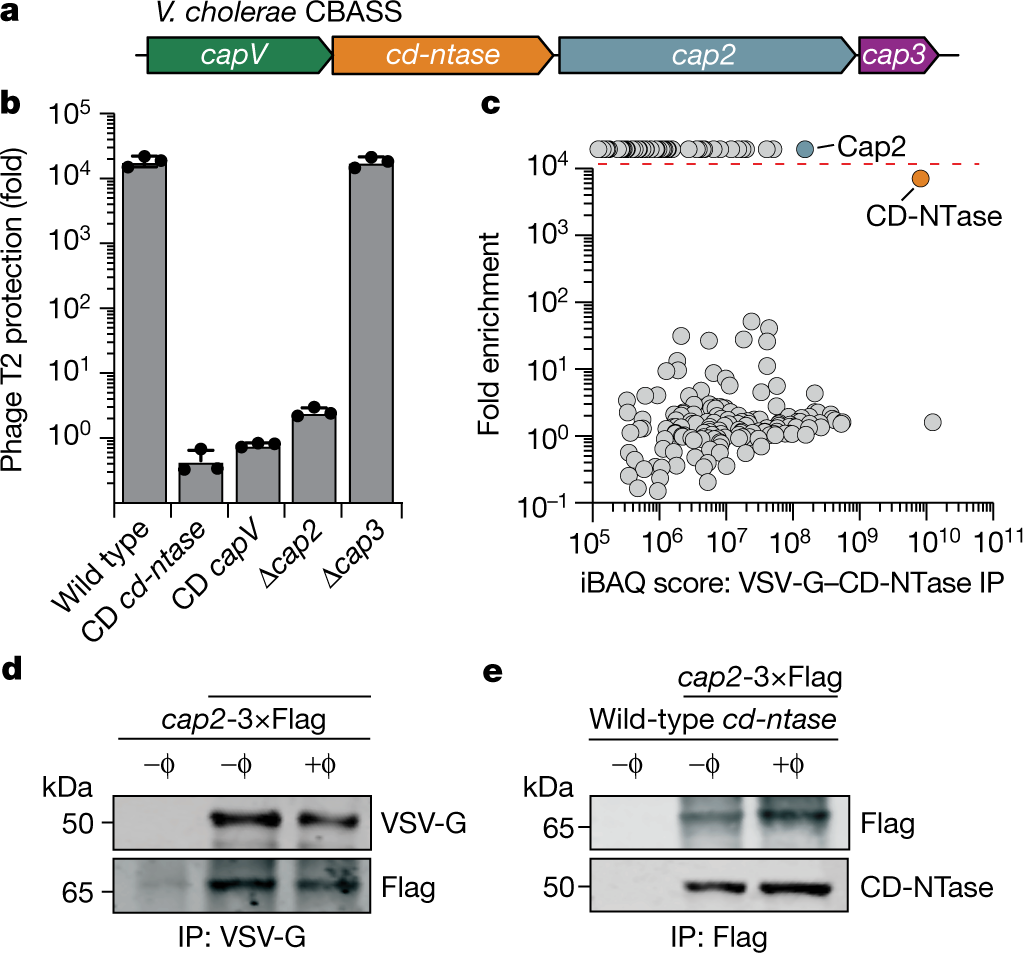
New carbohydrate binding domains identified by phage display based functional metagenomic screens of human gut microbiota | Communications Biology

TR0068-Protein-assay-compatibility.pdf - TECH TIP #68 Protein assay compatibility table TR0068.5 Introduction No single protein assay method is | Course Hero

Proteotoxicity caused by perturbed protein complexes underlies hybrid incompatibility in yeast | Nature Communications

TR0068-Protein-assay-compatibility.pdf - TECH TIP #68 Protein assay compatibility table TR0068.5 Introduction No single protein assay method is | Course Hero
Biomolecules | Free Full-Text | Recent Progress in Development and Application of DNA, Protein, Peptide, Glycan, Antibody, and Aptamer Microarrays

TR0068-Protein-assay-compatibility.pdf - TECH TIP #68 Protein assay compatibility table TR0068.5 Introduction No single protein assay method is | Course Hero
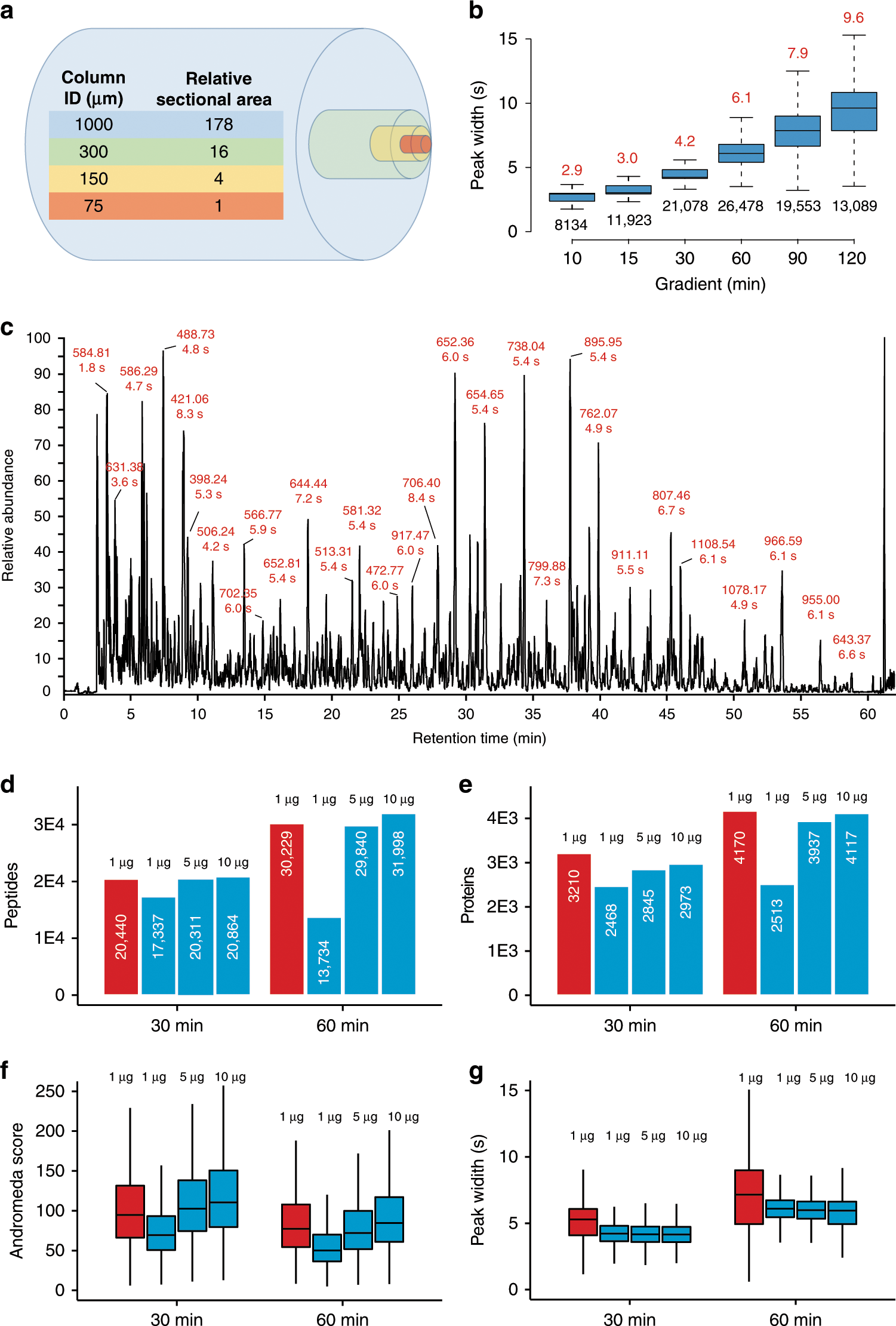
Robust, reproducible and quantitative analysis of thousands of proteomes by micro-flow LC–MS/MS | Nature Communications
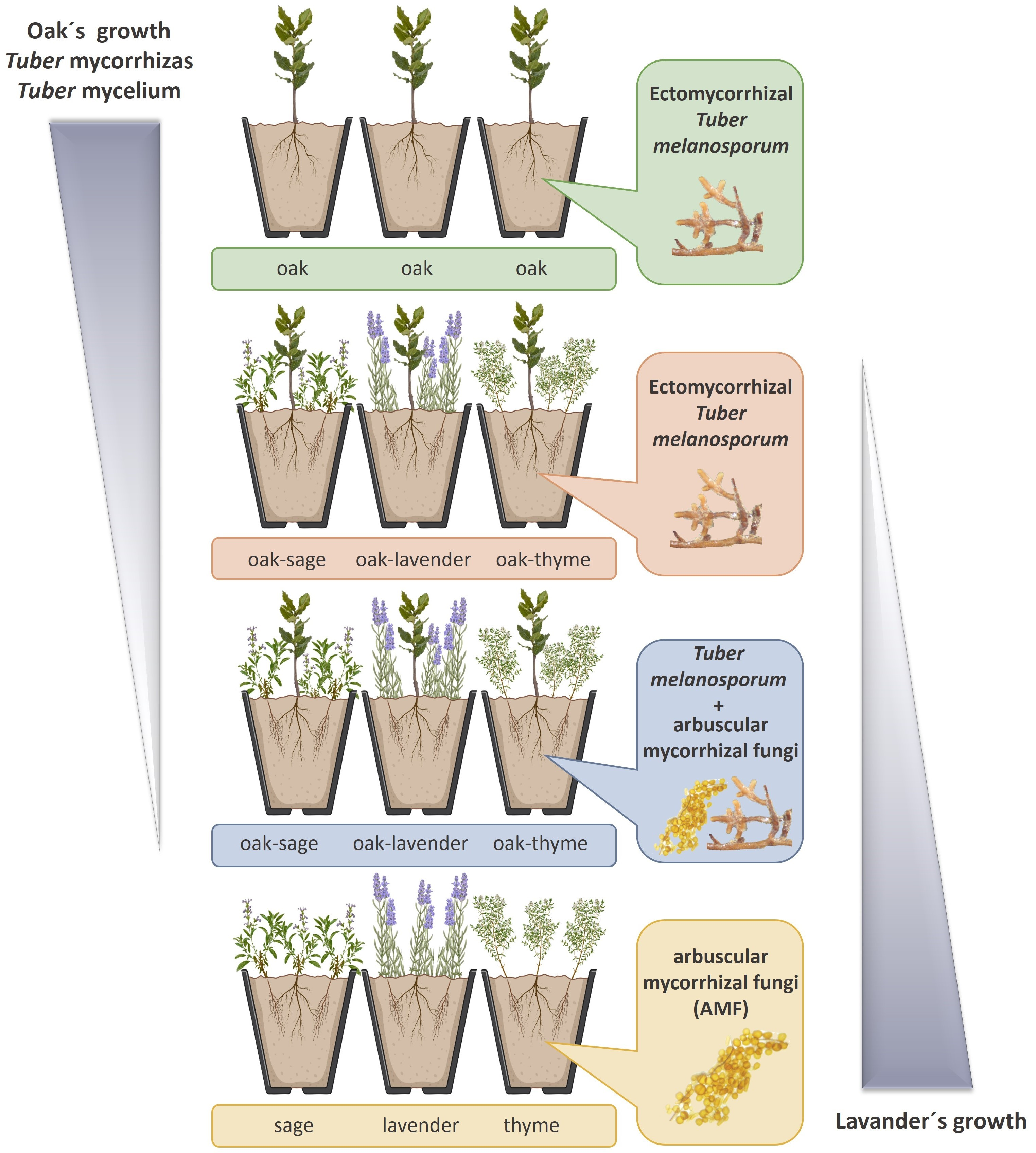
Biology | Free Full-Text | Aromatic Plants and Their Associated Arbuscular Mycorrhizal Fungi Outcompete Tuber melanosporum in Compatibility Assays with Truffle-Oaks

TR0068-Protein-assay-compatibility.pdf - TECH TIP #68 Protein assay compatibility table TR0068.5 Introduction No single protein assay method is | Course Hero

Quantitative Profiling of WNT-3A Binding to All Human Frizzled Paralogues in HEK293 Cells by NanoBiT/BRET Assessments | ACS Pharmacology & Translational Science

TR0068-Protein-assay-compatibility.pdf - TECH TIP #68 Protein assay compatibility table TR0068.5 Introduction No single protein assay method is | Course Hero

Assessment of Label-Free Quantification in Discovery Proteomics and Impact of Technological Factors and Natural Variability of Protein Abundance | Journal of Proteome Research

TR0068-Protein-assay-compatibility.pdf - TECH TIP #68 Protein assay compatibility table TR0068.5 Introduction No single protein assay method is | Course Hero
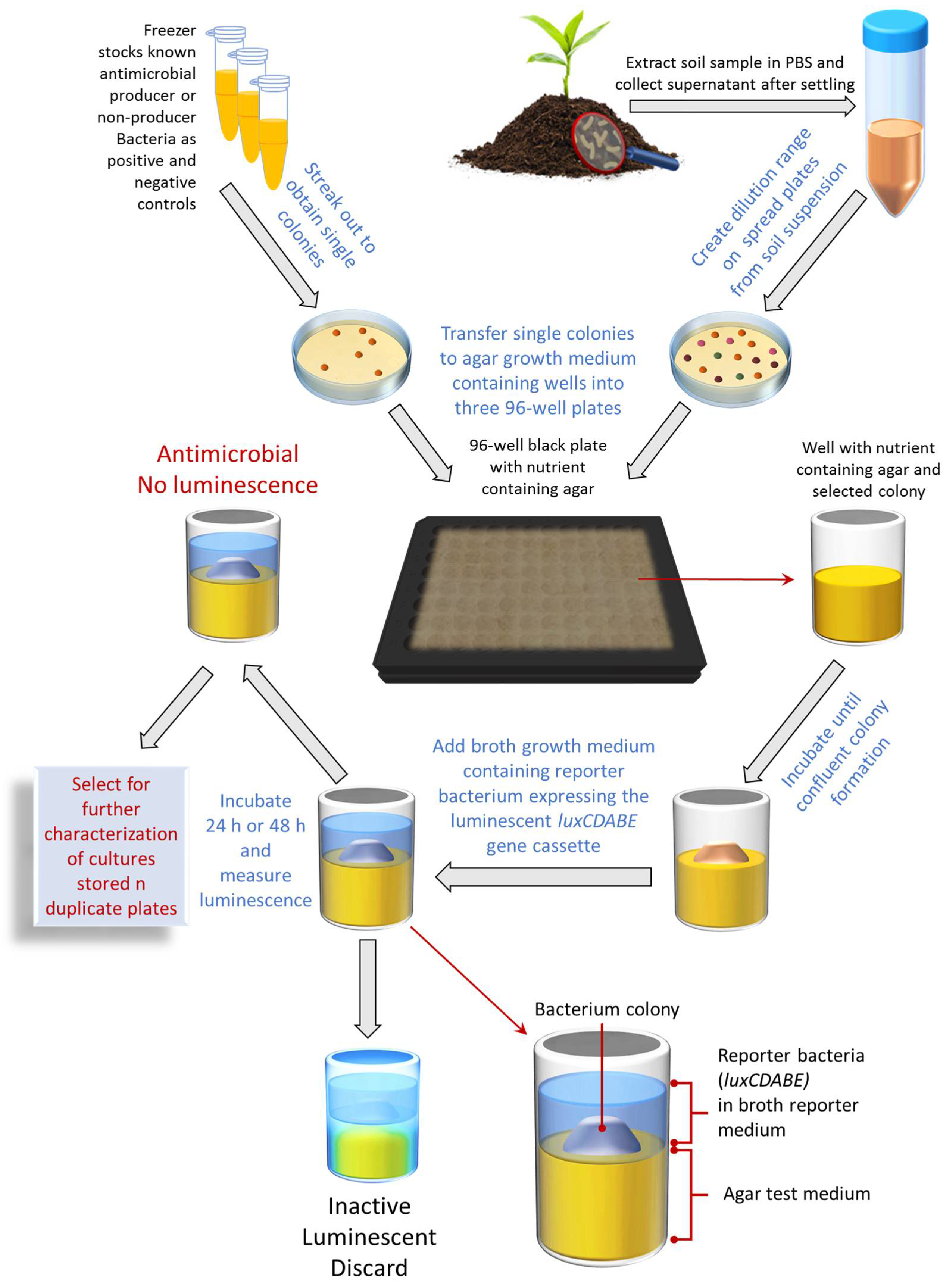
Microorganisms | Free Full-Text | Direct Detection of Antibacterial-Producing Soil Isolates Utilizing a Novel High-Throughput Screening Assay

Fillable Online Protein assay compatibility table - Thermo Fisher Scientific Fax Email Print - pdfFiller

LC–MS-based quantification of intact proteins: perspective for clinical and bioanalytical applications | Bioanalysis